Understanding the Complexity of Genetic Alzheimer’s Disease
By Sarah J. Eger
In our recent study, we delved into the cellular differences between two forms of Alzheimer’s Disease (AD).
To uncover their distinct characteristics, we focused on autosomal dominant Alzheimer’s Disease (ADAD), which involves a highly penetrant genetic mutation. All ADAD cases in our study carried the PSEN1 E280A mutation, and these individuals are part of the largest known kindred in the world, located in Colombia. This unique population sheds light on the genetic underpinnings of AD.
In contrast, sporadic AD, the more common form, results from a combination of genetic and environmental factors, and its complexity accounts for the majority of AD cases worldwide. What is common between the two is that upon neuropathological evaluation, cases will exhibit amyloid plaques and neurofibrillary tangles (NFTs) of TAU proteins.
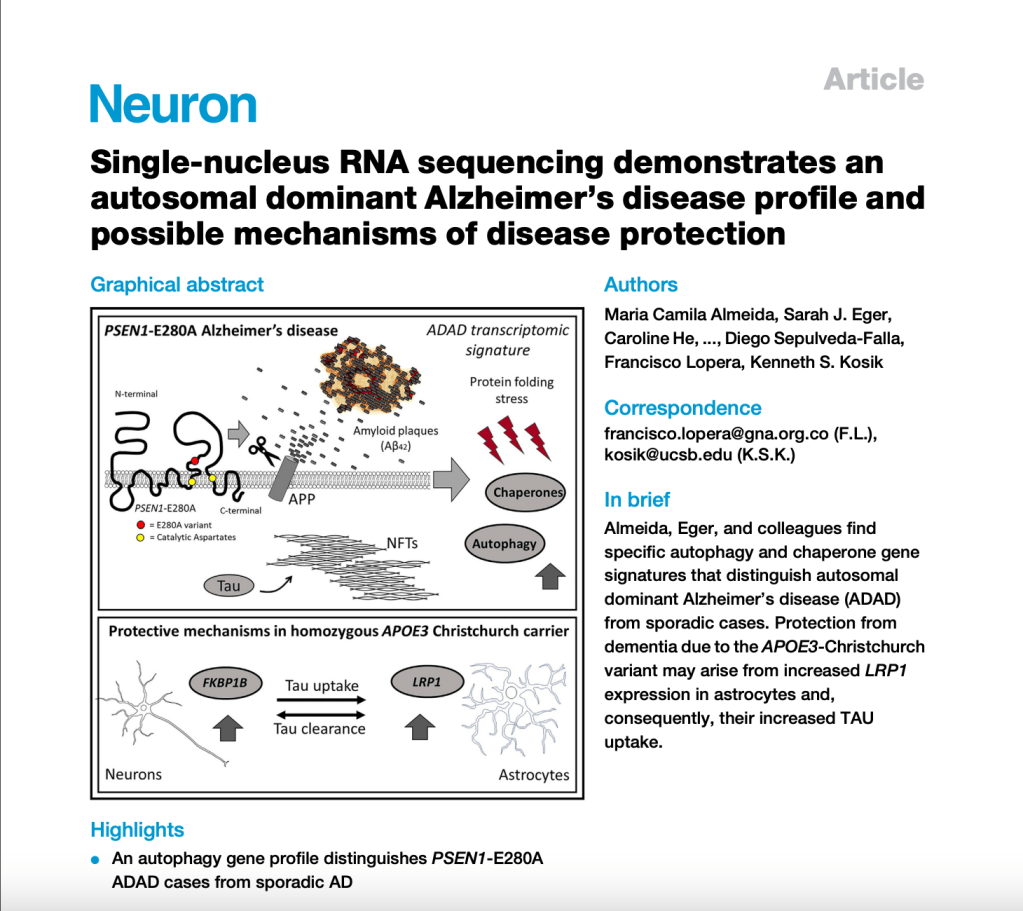
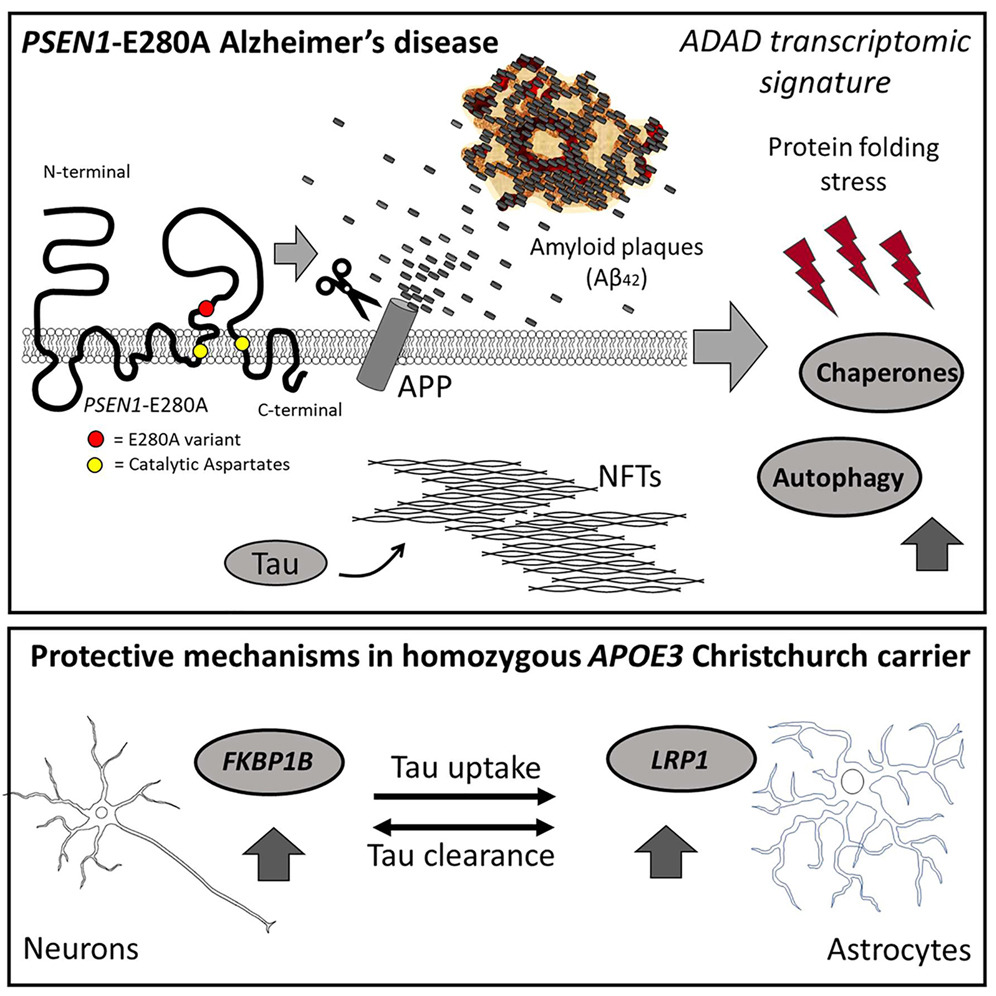
Analyzing gene activity at the single-cell level in brain tissue samples from ADAD, sporadic AD, and healthy controls, we found intriguing differences. ADAD cases showed significant upregulation of autophagy-related genes and chaperones, suggesting distinct biological pathways that involve increased protein folding stress, compared to sporadic AD.
Moreover, our study revealed potential mechanisms of neuroprotection in a particularly fascinating case within ADAD—a homozygous carrier of the rare APOE3 Christchurch variant. This individual displayed minimal neurofibrillary pathology and had remained cognitively normal decades after the average age at disease onset.
LRP1 was found to be upregulated in astrocytes of this individual compared to the other ADAD cases. It is known to play a crucial role in mediating the uptake and spread of TAU protein in neurons, which is central to the progression of AD pathology. Additionally, FKBP1B was upregulated in neurons for the homozygous carrier. This gene and others in its family have been shown to have an ameliorative effect on TAU inclusions, offering further insights into potential therapeutic targets for AD. These findings highlight the complexity of Alzheimer’s Disease and emphasize the importance of considering genetic factors when developing treatments and clinical trials. By understanding the unique cellular responses in different forms of the disease, we can tailor interventions to target specific pathways, offering hope for more effective therapies in the future.
Almeida, M. C., Eger, S. J., He, C., Audouard, M., Nikitina, A., Glasauer, S. M. K., Han, D., Mejía-Cupajita, B., Acosta-Uribe, J., Villalba-Moreno, N. D., Littau, J. L., Elcheikhali, M., Rivera, E. K., Carrettiero, D. C., Villegas-Lanau, C. A., Sepulveda-Falla, D., Lopera, F., & Kosik, K. S. (2024). Single-nucleus RNA sequencing demonstrates an autosomal dominant alzheimer’s disease profile and possible mechanisms of disease protection. Neuron, 112(11). https://doi.org/10.1016/j.neuron.2024.02.009